Multinomial Logistic Regression
Nov 13, 2023
Announcements
Due dates
- Draft report due in GitHub repo on 9am on your lab day
- HW 04 due TODAY at 11:59pm
- Statistics experience due Mon, Nov 20 at 11:59pm
Next week:
- (Optional) Project meetings Nov 20 & 21. Click here to sign up. Must sign up by Fri, Nov 17
- No class meetings next week
Click here to access lecture recordings. Available until Mon, Dec 04 at 9am
Topics
Conditions for logistic regression AE
Multinomial logistic regression
Interpret model coefficients
Inference for a coefficient \(\beta_{jk}\)
Application exercise
Computational setup
Generalized Linear Models
Generalized Linear Models (GLMs)
In practice, there are many different types of outcome variables:
- Binary: Win or Lose
- Nominal: Democrat, Republican or Third Party candidate
- Ordered: Movie rating (1 - 5 stars)
- and others…
Predicting each of these outcomes requires a generalized linear model, a broader class of models that generalize the multiple linear regression model
Note
Recommended reading for more details about GLMs: Generalized Linear Models: A Unifying Theory.
Binary outcome (Logistic)
Given \(P(y_i=1|x_i)= \hat{\pi}_i\hspace{5mm} \text{ and } \hspace{5mm}P(y_i=0|x_i) = 1-\hat{\pi}_i\)
\[ \log\Big(\frac{\hat{\pi}_i}{1-\hat{\pi}_i}\Big) = \hat{\beta}_0 + \hat{\beta}_1 x_{i} \]
We can calculate \(\hat{\pi}_i\) by solving the logit equation:
\[ \hat{\pi}_i = \frac{\exp\{\hat{\beta}_0 + \hat{\beta}_1 x_{i}\}}{1 + \exp\{\hat{\beta}_0 + \hat{\beta}_1 x_{i}\}} \]
Binary outcome (Logistic)
Suppose we consider \(y=0\) the baseline category such that
\[ P(y_i=1|x_i) = \hat{\pi}_{i1} \hspace{2mm} \text{ and } \hspace{2mm} P(y_i=0|x_i) = \hat{\pi}_{i0} \]
Then the logistic regression model is
\[ \log\bigg(\frac{\hat{\pi}_{i1}}{1- \hat{\pi}_{i1}}\bigg) = \log\bigg(\frac{\hat{\pi}_{i1}}{\hat{\pi}_{i0}}\bigg) = \hat{\beta}_0 + \hat{\beta}_1 x_i \]
Slope, \(\hat{\beta}_1\): When \(x\) increases by one unit, the odds of \(y=1\) versus the baseline \(y=0\) are expected to multiply by a factor of \(\exp\{\hat{\beta}_1\}\)
Intercept, \(\hat{\beta}_0\): When \(x=0\), the predicted odds of \(y=1\) versus the baseline \(y=0\) are \(\exp\{\hat{\beta}_0\}\)
Multinomial outcome variable
Suppose the outcome variable \(y\) is categorical and can take values \(1, 2, \ldots, K\) such that \((K > 2)\)
Multinomial Distribution:
\[ P(y=1) = \pi_1, P(y=2) = \pi_2, \ldots, P(y=K) = \pi_K \]
such that \(\sum\limits_{k=1}^{K} \pi_k = 1\)
Multinomial Logistic Regression
If we have an explanatory variable \(x\), then we want to fit a model such that \(P(y = k) = \pi_k\) is a function of \(x\)
Choose a baseline category. Let’s choose \(y=1\). Then,
\[ \log\bigg(\frac{\pi_{ik}}{\pi_{i1}}\bigg) = \beta_{0k} + \beta_{1k} x_i \]
In the multinomial logistic model, we have a separate equation for each category of the outcome relative to the baseline category
- If the outcome has \(K\) possible categories, there will be \(K-1\) equations as part of the multinomial logistic model
Multinomial Logistic Regression
Suppose we have a outcome variable \(y\) that can take three possible outcomes that are coded as “A”, “B”, “C”
Let “A” be the baseline category. Then
\[ \begin{aligned} \log\bigg(\frac{\pi_{iB}}{\pi_{iA}}\bigg) &= \beta_{0B} + \beta_{1B}x_i \\[10pt] \log\bigg(\frac{\pi_{iC}}{\pi_{iA}}\bigg) &= \beta_{0C} + \beta_{1C} x_i \end{aligned} \]
Data
NHANES Data
National Health and Nutrition Examination Survey is conducted by the National Center for Health Statistics (NCHS)
The goal is to “assess the health and nutritional status of adults and children in the United States”
This survey includes an interview and a physical examination
NHANES Data
We will use the data from the
NHANESR packageContains 75 variables for the 2009 - 2010 and 2011 - 2012 sample years
The data in this package is modified for educational purposes and should not be used for research
Original data can be obtained from the NCHS website for research purposes
Type
?NHANESin console to see list of variables and definitions
Variables
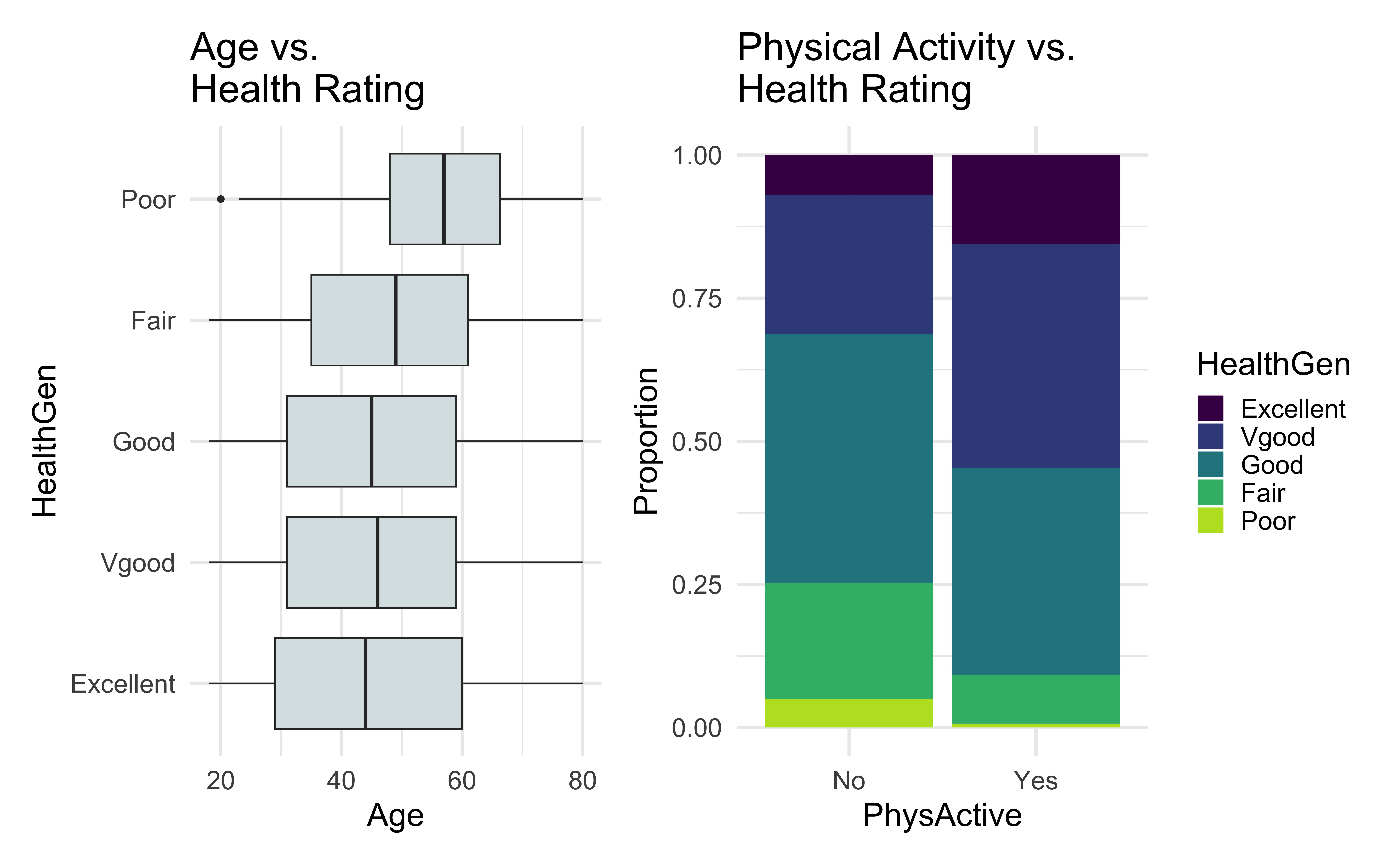
Goal: Use a person’s age and whether they do regular physical activity to predict their self-reported health rating.
Outcome:
HealthGen: Self-reported rating of participant’s health in general. Excellent, Vgood, Good, Fair, or Poor.
Predictors:
Age: Age at time of screening (in years). Participants 80 or older were recorded as 80.PhysActive: Participant does moderate to vigorous-intensity sports, fitness or recreational activities.
The data
Rows: 6,710
Columns: 4
$ HealthGen <fct> Good, Good, Good, Good, Vgood, Vgood, Vgood, Vgood, Vgood, …
$ Age <int> 34, 34, 34, 49, 45, 45, 45, 66, 58, 54, 50, 33, 60, 56, 56,…
$ PhysActive <fct> No, No, No, No, Yes, Yes, Yes, Yes, Yes, Yes, Yes, No, No, …
$ obs_num <int> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, …Exploratory data analysis

Exploratory data analysis
Fitting a multinomial logistic regression model
Model in R
Use the multinom_reg() function with the "nnet" engine:
Model result
parsnip model object
Call:
nnet::multinom(formula = HealthGen ~ Age + PhysActive, data = data,
trace = FALSE)
Coefficients:
(Intercept) Age PhysActiveYes
Vgood 1.2053460 0.0009101848 -0.3209047
Good 1.9476261 -0.0023686122 -1.0014925
Fair 0.9145492 0.0030462534 -1.6454297
Poor -1.5211414 0.0221905681 -2.6556343
Residual Deviance: 17588.88
AIC: 17612.88 Model output
What function do we use to get the model summary, i.e., coefficient estimates.
# A tibble: 12 × 6
y.level term estimate std.error statistic p.value
<chr> <chr> <dbl> <dbl> <dbl> <dbl>
1 Vgood (Intercept) 1.21 0.145 8.33 8.42e-17
2 Vgood Age 0.000910 0.00246 0.369 7.12e- 1
3 Vgood PhysActiveYes -0.321 0.0929 -3.45 5.51e- 4
4 Good (Intercept) 1.95 0.141 13.8 1.39e-43
5 Good Age -0.00237 0.00242 -0.977 3.29e- 1
6 Good PhysActiveYes -1.00 0.0901 -11.1 1.00e-28
7 Fair (Intercept) 0.915 0.164 5.57 2.61e- 8
8 Fair Age 0.00305 0.00288 1.06 2.90e- 1
9 Fair PhysActiveYes -1.65 0.107 -15.3 5.69e-53
10 Poor (Intercept) -1.52 0.290 -5.24 1.62e- 7
11 Poor Age 0.0222 0.00491 4.52 6.11e- 6
12 Poor PhysActiveYes -2.66 0.236 -11.3 1.75e-29Model output, with CI
# A tibble: 12 × 8
y.level term estimate std.error statistic p.value conf.low conf.high
<chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 Vgood (Intercept) 1.21e+0 0.145 8.33 8.42e-17 0.922 1.49
2 Vgood Age 9.10e-4 0.00246 0.369 7.12e- 1 -0.00392 0.00574
3 Vgood PhysActiveY… -3.21e-1 0.0929 -3.45 5.51e- 4 -0.503 -0.139
4 Good (Intercept) 1.95e+0 0.141 13.8 1.39e-43 1.67 2.22
5 Good Age -2.37e-3 0.00242 -0.977 3.29e- 1 -0.00712 0.00238
6 Good PhysActiveY… -1.00e+0 0.0901 -11.1 1.00e-28 -1.18 -0.825
7 Fair (Intercept) 9.15e-1 0.164 5.57 2.61e- 8 0.592 1.24
8 Fair Age 3.05e-3 0.00288 1.06 2.90e- 1 -0.00260 0.00869
9 Fair PhysActiveY… -1.65e+0 0.107 -15.3 5.69e-53 -1.86 -1.43
10 Poor (Intercept) -1.52e+0 0.290 -5.24 1.62e- 7 -2.09 -0.952
11 Poor Age 2.22e-2 0.00491 4.52 6.11e- 6 0.0126 0.0318
12 Poor PhysActiveY… -2.66e+0 0.236 -11.3 1.75e-29 -3.12 -2.19 Model output, with CI
| y.level | term | estimate | std.error | statistic | p.value | conf.low | conf.high |
|---|---|---|---|---|---|---|---|
| Vgood | (Intercept) | 1.205 | 0.145 | 8.325 | 0.000 | 0.922 | 1.489 |
| Vgood | Age | 0.001 | 0.002 | 0.369 | 0.712 | -0.004 | 0.006 |
| Vgood | PhysActiveYes | -0.321 | 0.093 | -3.454 | 0.001 | -0.503 | -0.139 |
| Good | (Intercept) | 1.948 | 0.141 | 13.844 | 0.000 | 1.672 | 2.223 |
| Good | Age | -0.002 | 0.002 | -0.977 | 0.329 | -0.007 | 0.002 |
| Good | PhysActiveYes | -1.001 | 0.090 | -11.120 | 0.000 | -1.178 | -0.825 |
| Fair | (Intercept) | 0.915 | 0.164 | 5.566 | 0.000 | 0.592 | 1.237 |
| Fair | Age | 0.003 | 0.003 | 1.058 | 0.290 | -0.003 | 0.009 |
| Fair | PhysActiveYes | -1.645 | 0.107 | -15.319 | 0.000 | -1.856 | -1.435 |
| Poor | (Intercept) | -1.521 | 0.290 | -5.238 | 0.000 | -2.090 | -0.952 |
| Poor | Age | 0.022 | 0.005 | 4.522 | 0.000 | 0.013 | 0.032 |
| Poor | PhysActiveYes | -2.656 | 0.236 | -11.275 | 0.000 | -3.117 | -2.194 |
Fair vs. Excellent Health
The baseline category for the model is Excellent.
The model equation for the log-odds a person rates themselves as having “Fair” health vs. “Excellent” is
\[ \log\Big(\frac{\hat{\pi}_{Fair}}{\hat{\pi}_{Excellent}}\Big) = 0.915 + 0.003 ~ \text{age} - 1.645 ~ \text{PhysActive} \]
Interpretations
\[ \log\Big(\frac{\hat{\pi}_{Fair}}{\hat{\pi}_{Excellent}}\Big) = 0.915 + 0.003 ~ \text{age} - 1.645 ~ \text{PhysActive} \]
For each additional year in age, the odds a person rates themselves as having fair health versus excellent health are expected to multiply by 1.003 (exp(0.003)), holding physical activity constant.
The odds a person who does physical activity will rate themselves as having fair health versus excellent health are expected to be 0.193 (exp(-1.645)) times the odds for a person who doesn’t do physical activity, holding age constant.
Interpretations
\[ \log\Big(\frac{\hat{\pi}_{Fair}}{\hat{\pi}_{Excellent}}\Big) = 0.915 + 0.003 ~ \text{age} - 1.645 ~ \text{PhysActive} \]
The odds a 0 year old person who doesn’t do physical activity rates themselves as having fair health vs. excellent health are 2.497 (exp(0.915)).
Warning
Need to mean-center age for the intercept to have a meaningful interpretation!
Hypothesis test for \(\beta_{jk}\)
The test of significance for the coefficient \(\beta_{jk}\) is
Hypotheses: \(H_0: \beta_{jk} = 0 \hspace{2mm} \text{ vs } \hspace{2mm} H_a: \beta_{jk} \neq 0\)
Test Statistic: \[z = \frac{\hat{\beta}_{jk} - 0}{SE(\hat{\beta_{jk}})}\]
P-value: \(P(|Z| > |z|)\),
where \(Z \sim N(0, 1)\), the Standard Normal distribution
Confidence interval for \(\beta_{jk}\)
We can calculate the C% confidence interval for \(\beta_{jk}\) using \(\hat{\beta}_{jk} \pm z^* SE(\hat{\beta}_{jk})\), where \(z^*\) is calculated from the \(N(0,1)\) distribution.
Interpretation: We are \(C\%\) confident that for every one unit change in \(x_{j}\), the odds of \(y = k\) versus the baseline will multiply by a factor of \(\exp\{\hat{\beta}_{jk} - z^* SE(\hat{\beta}_{jk})\}\) to \(\exp\{\hat{\beta}_{jk} + z^* SE(\hat{\beta}_{jk})\}\), holding all else constant.
Interpreting CIs for \(\beta_{jk}\)
| y.level | term | estimate | std.error | statistic | p.value | conf.low | conf.high |
|---|---|---|---|---|---|---|---|
| Fair | (Intercept) | 0.915 | 0.164 | 5.566 | 0.00 | 0.592 | 1.237 |
| Fair | Age | 0.003 | 0.003 | 1.058 | 0.29 | -0.003 | 0.009 |
| Fair | PhysActiveYes | -1.645 | 0.107 | -15.319 | 0.00 | -1.856 | -1.435 |
We are 95% confident, that for each additional year in age, the odds a person rates themselves as having fair health versus excellent health will multiply by 0.997 (exp(-0.003)) to 1.009 (exp(0.009)) , holding physical activity constant.
Interpreting CIs for \(\beta_{jk}\)
| y.level | term | estimate | std.error | statistic | p.value | conf.low | conf.high |
|---|---|---|---|---|---|---|---|
| Fair | (Intercept) | 0.915 | 0.164 | 5.566 | 0.00 | 0.592 | 1.237 |
| Fair | Age | 0.003 | 0.003 | 1.058 | 0.29 | -0.003 | 0.009 |
| Fair | PhysActiveYes | -1.645 | 0.107 | -15.319 | 0.00 | -1.856 | -1.435 |
We are 95% confident that the odds a person who does physical activity will rate themselves as having fair health versus excellent health are 0.156 (exp(-1.856 )) to 0.238 (exp(-1.435)) times the odds for a person who doesn’t do physical activity, holding age constant.
Recap
Introduce multinomial logistic regression
Interpret model coefficients
Inference for a coefficient \(\beta_{jk}\)